3.1 透過GBIF發布生物多樣性資料集
資料集包括了詮釋資料和原始資料。最簡單的資料集可以只包含詮釋資料。目前GBIF支持發布的資料集有以下四種:1) 僅詮釋資料 metadata-only,2) 物種名錄 cheklist,3) 出現紀錄 occurrence,以及4) 調查活動資料集 sampling event,依照資料內容的豐富性及資料結構的複雜性而定。 為了維持一個資料集的完整性,我們建議資料提供者盡可能發布現有最完整的資料集。
在網站上發布資料集就像是生產資料產品;而詮釋資料就像產品專屬的使用手冊,用來告訴使用者產品的內容、產品的使用方法以及客服相關資訊等。換句話說,詮釋資料可以告訴你資料的來源、如何使用該資料,以及如果對資料有任何疑問時可以聯繫誰等。一個產品就是一種資源,因此,資料集也是網路的資源。在GBIF的概念中,資料集是一種資源,要透過GBIF發布資料集(或資源),必須使用符合達爾文核心延伸集中一連串的專有名詞作為資料欄位標準名稱,包含物種名錄、出現紀錄、調查活動資料集的資訊都要按照此標準呈現。
準備資料集最簡單的方法是使用預定格式的Excel模板來填寫詮釋資料和原始資料。這將協助你輕鬆地將資料整理後輸入到GBIF接受的標準化資料結構中。GBIF提供簡單的Excel模板,其中包括達爾文核心集所採用的標準化資料欄位名稱。
但是,當你的資料表中有大量的資料時(例如超過一萬筆以上的紀錄),你會發現使用Excel處理這些紀錄很困難。在這種情況下,我們建議可以使用R,R是一種提供統計和資料處理的開源軟體。如果你是R的新手,我們鼓勵你學習使用R,因為它是目前最受歡迎的資料分析工具之一。在本章節中,我們將向你說明如何使用Excel模板和R script來準備調查活動資料集。
3.2 什麼是調查活動資料?
有時資料集可提供的資訊遠超乎你想像,不僅僅可提供發現物種的地點、日期,也有機會去觸及更廣泛的物種分類群的群落組成或者不同時間、地點的物種豐度。這些資料集通常來自測量和監測生物多樣性的標準作業程序,如植物群落穿越線採樣調查、鳥類調查計畫以及淡水和海水採樣。藉由指出記錄樣本時所使用的方法、事件和物種相對豐度,這些資料集可以強化在同樣標準作業程序下,但在不同時間不同地點所收集的資料之間的比較,有時候甚至可以讓研究員發現某些特定地點物種缺失的狀況。 (原文參照:http://www.gbif.org/publishing-data/summary)
3.2.1 調查活動(Event)+ 位置(Location)之核心(Core)詞彙
(**為必填欄位)| 調查活動識別碼(eventID**) | 最低海拔(公尺)(minimumElevationInMeters) |
| 父階調查活動識別碼(parentEventID) | 最高海拔(公尺) (maximumElevationInMeters) |
| 調查活動日期(eventDate**) | 最小深度(公尺) (minimumDepthInMeters) |
| 棲息地(Habitat) | 最大深度(公尺) (maximumDepthInMeters) |
| 調查採樣流程(samplingProtocol**) | 與海平面最小距離(公尺) (minimumDistanceAboveSurfaceIn Meters) |
| 樣本大小值(sampleSizeValue**) | 與海平面最大距離(公尺) (maximumDistanceAboveSurfaceIn Meters) |
| 樣本大小單位(sampleSizeUnit**) | 洲(Continent) |
| 調查採樣努力量(samplingEffort) | 國家(Country) |
| 地點識別碼(locationID) | 國碼(countryCode) |
| 十進位緯度(decimalLatitude) | 州/省(stateProvince) |
| 十進位經度(decimalLongitude) | 縣(Country) |
| 大地基準(geodeticDatum) | 市(Municipality) |
| 座標誤差(公尺)(coordinateUncertaintyInMeters*) | 地點(locality) |
| 座標精確度(coordinatePrecision*) |
參照:http://tools.gbif.org/dwca-validator/extension.do?id=dwc:Event
3.2.2 測量或事實(MeasurementOrFact)之核心詞彙
- 調查活動識別碼(eventID**)
- 測量識別碼(measurementID
- 測量類型(measurementType)
- 測量值(measurementValue)
- 測量準確度(measurementAccuracy)
- 測量單位(measurementUnit)
- 測量確認者(measurementDeterminedBy)
- 測量確認日期(measurementDeterminedDate)
- 測量方法(measurementMethod)
- 測量備註(measurementRemarks)
參照:http://tools.gbif.org/dwca-validator/extension.do?id=dwc:MeasurementOrFact
3.2.3 選擇出現紀錄(Occurrence)+ 分類(Taxon)之核心詞彙
| 調查活動識別碼(eventID**) | 個體數(individualCount) |
| 出現紀錄識別碼(occurrenceID**) | 生物體數量(organismQuantity) |
| 學名識別碼(scientificNameID) | 生物體數量類別(organismQuantityType) |
| 學名(scientificName) | 性別(sex) |
| 分類識別碼(taxonID) | 生活史階段(lifestage) |
| 俗名(vernacularName) | 繁殖狀況(reproductiveCondition) |
| 分類階層(taxonRank) | 行為(behavior) |
| 界(kingdom) | 出現紀錄狀態(occurrenceStatus) |
| 門(phylum) | 有效學名使用名稱識別碼(acceptedNameUsageID) |
| 綱(class) | 有效學名使用名稱(acceptedNameUsage) |
| 目(order) | 種小名(specificEpithet) |
| 科(family) | 亞種小名(infraspecificEpithet) |
| 屬(genus) | 學名命名者(scientificNameAuthorship) |
| 記錄者(recordedBy) |
參照:http://tools.gbif.org/dwca-validator/extension.do?id=dwc:Occurrence
3.3 達爾文核心集—資料的星型結構

3.3.1 資料的星型結構 (Star schema)
GBIF 的發布資料結構如上圖所示(以調查活動為例),在發布資料前,先確認你的研究資料類型是屬於名錄、出現紀錄還是調查活動,並以該類型資料結構為基礎(Core核心集)去設定資料表的欄位名稱(參照 Darwin Core),而其他未被包含於核心集的資料,則可以選擇其他類型的欄位名稱,整理成延伸集(可視為核心集的補充說明)。所有的延伸集都必須能有一個欄位(強烈建議為唯一識別碼ID)與核心集相同,才能相互對應,追溯到同一批次的資料。可以把這些想像成一個excel表,主頁資料就是核心集,其他分頁都是延伸集,內容皆與主頁有關。
3.3.2 如何發布資料呢?
- 先準備好原始資料
- 檢查並清理資料(修正學名或其他的錯誤)
- 採用達爾文核心調查活動、出現紀錄等詞彙集,作為資料表的欄位名稱
- 將資料結構轉成獨立的表格,並將資料套入達爾文核心星型結構中,區分核心集(Core)和延伸集(Extension),接著將此表格儲存為以UTF-8編碼的CSV文本檔案
- 準備詮釋資料
- 在由節點管理的IPT工具(如TaiBIF IPT)上申請帳號資料,節點將確認你的組織成為資料發布組織(須先於GBIF註冊申請成為資料發布者)
- 登入IPT,建立新的資料來源,上傳你的資料表並與Darwin Core進行欄位對應
- 提供詮釋資料
- 發布資料


如何發布調查活動資料?
http://github.com/gbif/ipt/wiki/samplingEventData#exemplar-datasets
大致流程:
確認資料的種類 >> 轉換資料 >> 上傳資料至IPT >> 將資料對應至調查活動及出現紀錄 >> 填入詮釋資料 >> 發布資料集 >> 完成資料集註冊
3.4 使用Excel模板準備調查活動資料
範例:準備資料所使用的資料集與工具
- 範例表格 1_AlienPlantSurvey.xlsx (範例:外來種植物調查)
- 試算表軟體 MS Excel / LibreOffice Calc
- 純文字編輯器 Notepad++ / PSPad / …
- 臺灣生物多樣性資訊機構學名比對工具 (http://match.taibif.tw)
範例:資料檔案
- 1_AlienPlantSurvey.xlsx (範例:外來種植物調查)
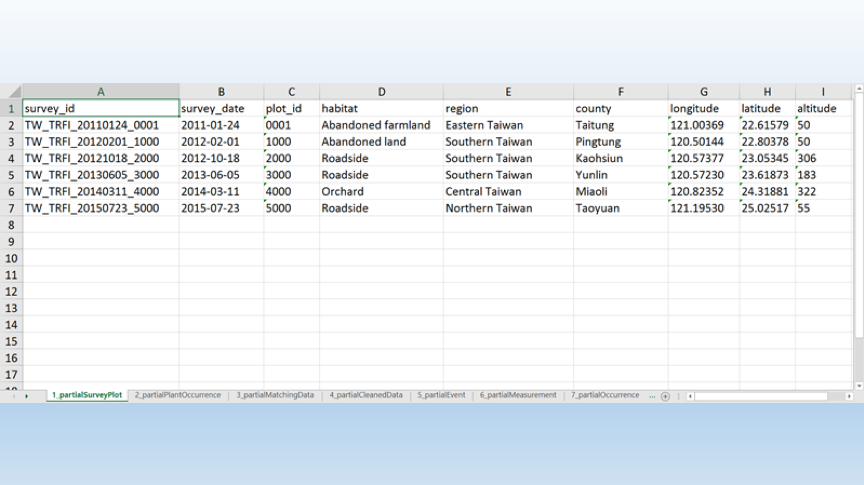
- 7個表格:
- 部分調查樣區(partialSurveyPlot)
- 部分植物出現紀錄(partialplantOccurrence)
- 部分已對應資料(partialMatchingData)
- 部分已清理資料(partialCleanedData)
- 部分調查活動(partialEvent)
- 部分測量值(partialMeasurement)
- 部分出現紀錄(partialOccurrence)

資料清理和轉換之工作流程:
- 檢查並研讀原始資料結構
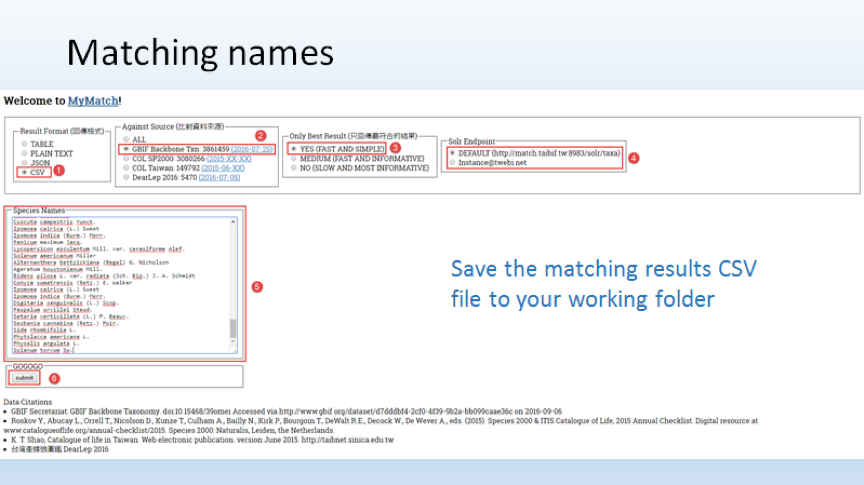
- 使用學名比對工具檢查輸入的學名拼法是否正確,並將配對結果儲存為CSV文字檔
- 使用NotePad ++打開上述文件,複製所有內容並貼到試算表
- 根據配對結果整理名稱,獲取分類階層的資訊以及更高分類階層名稱
- 分別創建調查活動、測量或事實以及出現紀錄的表格
- 將原始欄位名稱對應到達爾文核心詞彙
- 分別複製調查活動、測量、出現紀錄表格的資料到Notepad++,以UTF-8編碼,接著再存成CSV文字檔


 網站捷徑
網站捷徑





